CSNAP (Chemical Similarity Network Analysis Pull-down) Web
somthing
A network-based approach for automated compound target identification
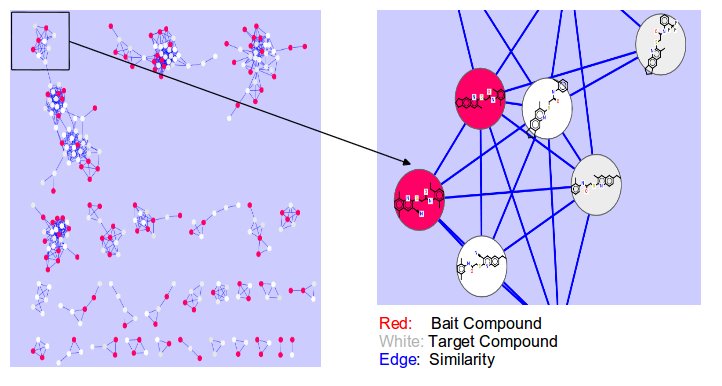
CSNAP (Chemical Similarity Network Analysis Pull-down) is a computational approach for compound target identification based on network similarity graphs. Query and reference compounds are populated on the network connectivity map and a graph-based neighbor counting method is applied to rank the consensus targets among the neighborhood of each query ligand. The CSNAP approach can facilitate high-throughput target discovery and off-target prediction for any compound set identified from phenotype-based or cell-based chemical screens.
CSNAP web server (version 1.0): start here The server is freely accessible without any log-in requirement. The website has been tested and optimized for Firefox, Opera, Chrome and Safari. Please see Q&A page for additional details.
Users please cite:
Support:
This material is based upon work supported by the National Science Foundation under Grant Number MCB1243645.