Disulfide bonds impart great conformation stability in proteins. They arise by the oxidation of the sulfhydryl (SH) groups of two cysteine amino acids linking two parts of the same protein chain or two separate chains together. As illustrated in the figure below, this linkage has five dihedral angles. Some combinations of these angles are more sterically favorable then others. This server calculates the dihedral angles of all disulfide bonds found in the uploaded structure PDB file. Based on these, a Dihedral Energy is calculated according to an empirical formula first defined by Katz et al. [Katz, BA, Kossiakoff A. J Biol Chem 1986 ; 261(33):15480-5.]. The Dihedral Energy, and dihedral angles from the uploaded are compared to disulfides found in a large non-redundant set of structures from the Protein Data Bank.
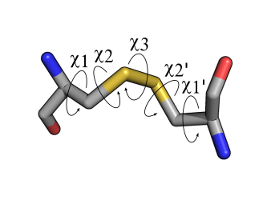 Model Disulfide Showing Five Dihedral Angles This is a stick representation
two disulfide bonded cystines, with atoms in CPK colors (i.e. carbon in grey,
oxygen in red, nitrogen in blue and sulfur in yellow). The assignment of the dihedral
angles are primed or unprimed emphasizing their interchangability depending on the
numbering of the disulfide bond.
The Dihedral Energy is calculated with the following equation:
E(kJ/mol)=8.37(1 + cos(3χ1)) + 8.37(1 + cos(3χ1')) + 4.18(1 + cos(3χ2))
+ 4.18(1 + cos(3χ2')) + 14.64(1 + cos(2χ3)) + 2.51(1 + cos(3χ3))
Model Disulfide Showing Five Dihedral Angles This is a stick representation
two disulfide bonded cystines, with atoms in CPK colors (i.e. carbon in grey,
oxygen in red, nitrogen in blue and sulfur in yellow). The assignment of the dihedral
angles are primed or unprimed emphasizing their interchangability depending on the
numbering of the disulfide bond.
The Dihedral Energy is calculated with the following equation:
E(kJ/mol)=8.37(1 + cos(3χ1)) + 8.37(1 + cos(3χ1')) + 4.18(1 + cos(3χ2))
+ 4.18(1 + cos(3χ2')) + 14.64(1 + cos(2χ3)) + 2.51(1 + cos(3χ3))
|