CSNAP (Chemical Similarity Network Analysis Pull-down) Web
Tutorial #1: Target Prediction and Design of HIV Inhibitors
- In the input field of the CSNAP query page, click "load example1", this should load the SMILES string of the HIV inhibitor in the text area.
- Accept the default search parameters then click "upload".
- A confirmation page of the input parameters will be created, click "submit".
- When the CSNAP analysis is completed, an output page will be created.
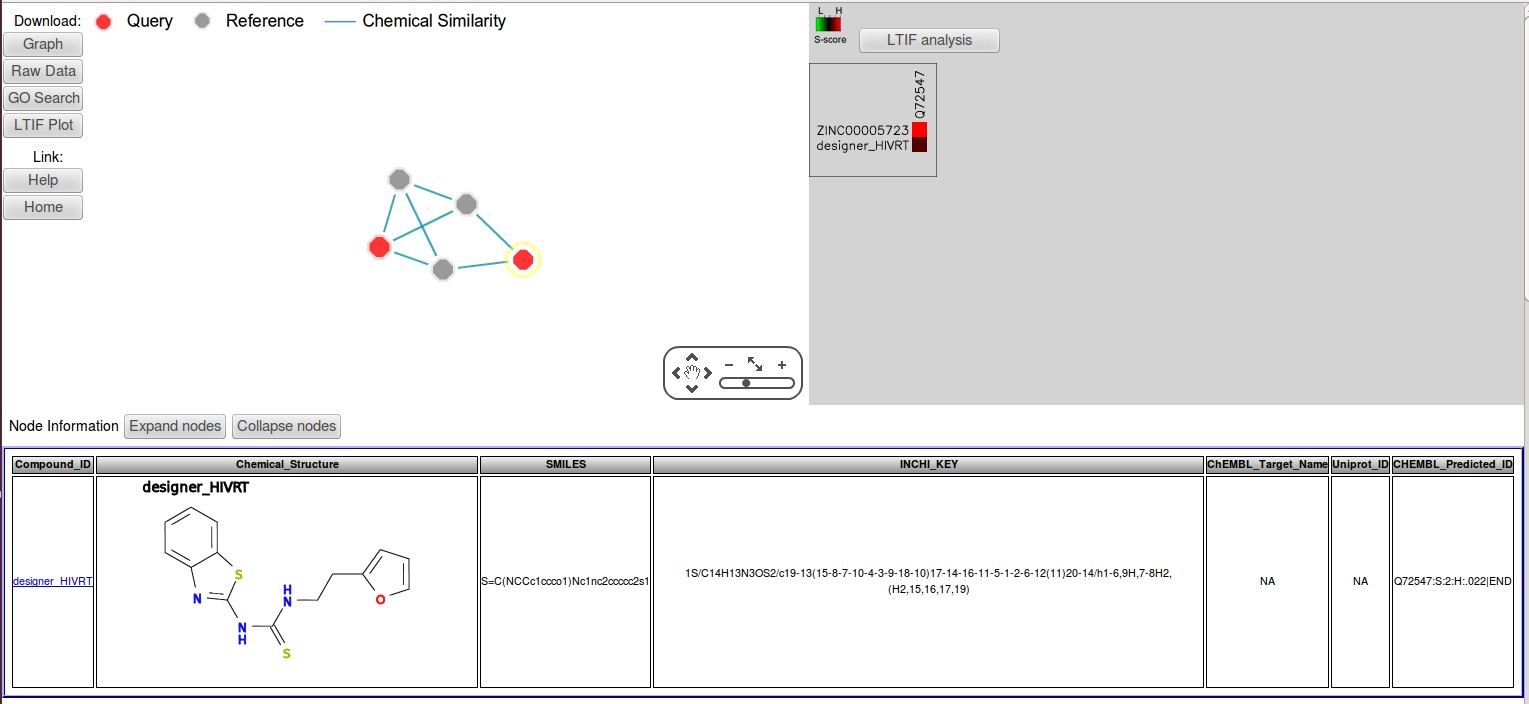
- To get more information about the output compounds, select both nodes in the network panel then click "Expand nodes" to enlarge the view area.
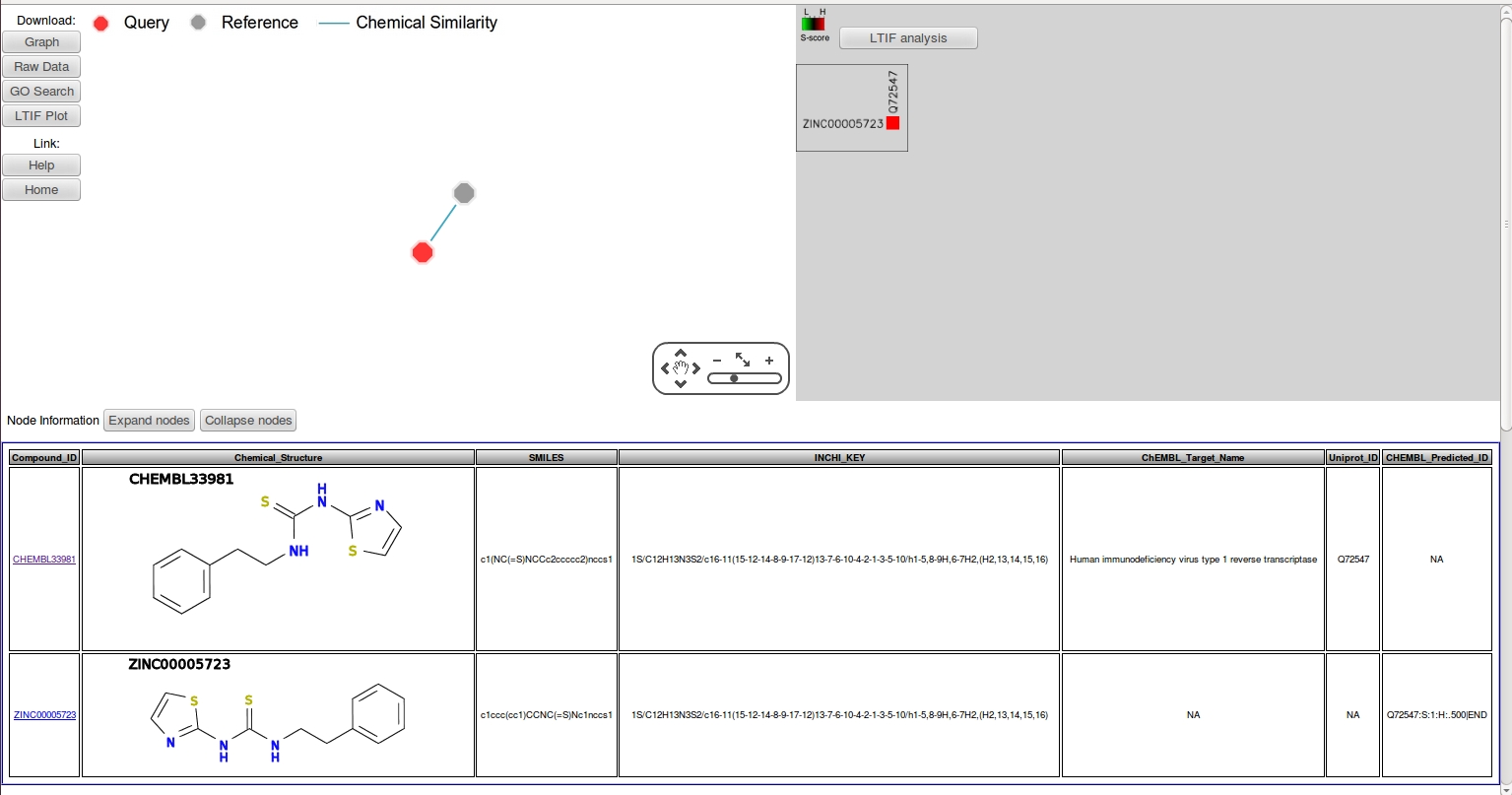
- The node information panel shows that both structures are identical. To confirm, click the edge connecting the two nodes, which will give a similarity value of "1" (identical) in the edge information panel. The "CHEMBL_target_name" column of the reference node, CHEMBL33981 indicates that the reference compound is an HIV reverse transcriptase (HIVRT) inhibitor. Thus, the query is predicted to be an HIVRT inhibitor with a S-score=1, where the target annotation was transferred directly from its immediate neighbor. Furthermore, the LTIF heatmap in the middle panel shows that the HIVRT inhibitor is highly specific with no predicted off-targets.
- To retrieve additional analogs for more in-depth SAR studies, apply different fingerprints for chemical similarity searches. First, return to CSNAP query page, click "load example1" to load the SMILES string of the HIVRT inhibitor we just analyzed.
- Adjust the default search fingerprint from "FP2" to "FP3", click "upload" then "submit" to proceed the calculation as previsouly described.
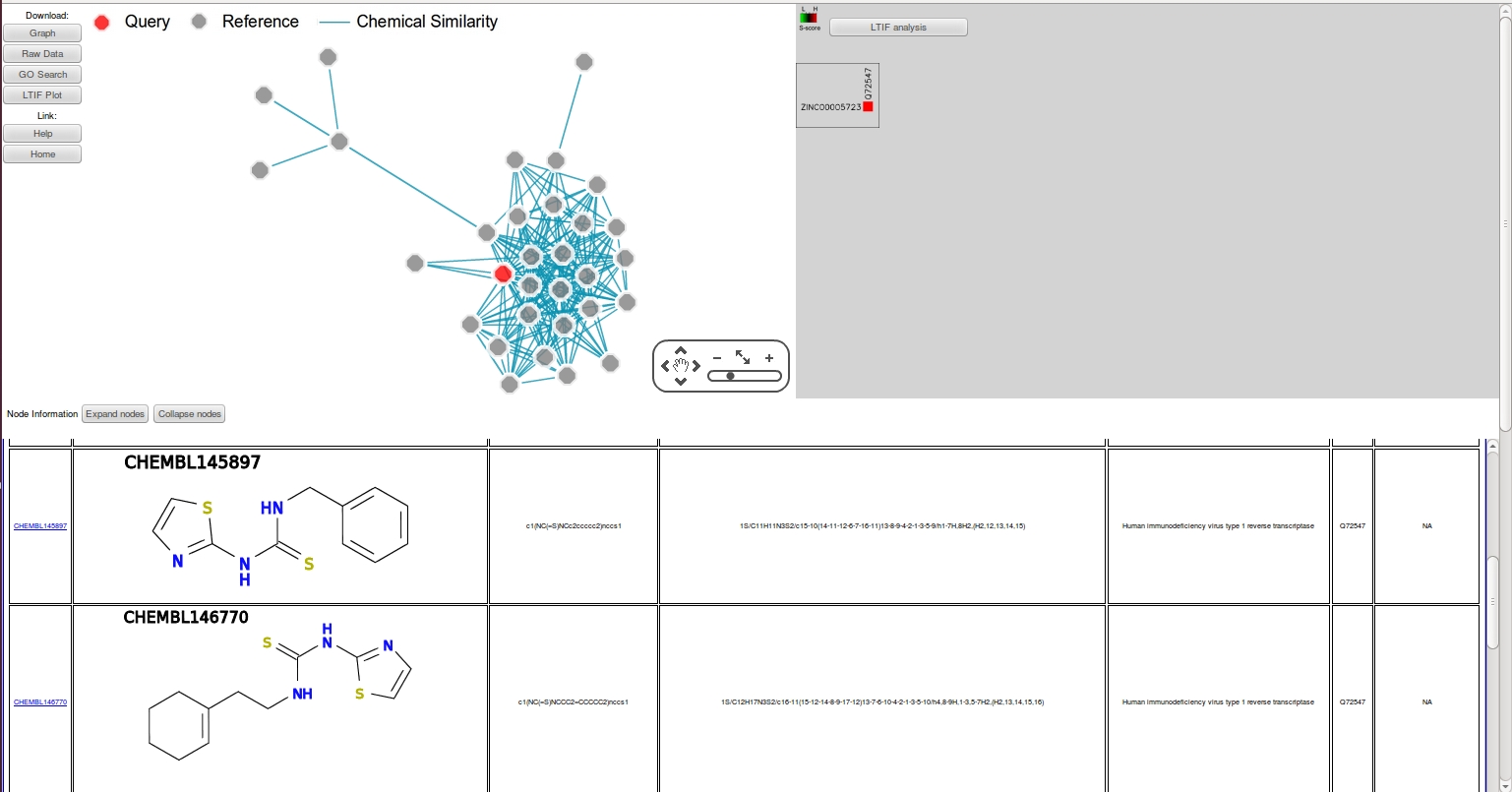
- Note that 31 additional HIVRT analogs have been retrieved this time and the S-score of the query has increased to 32. Furthermore, by analyzing the compound structures in the network, we have identified a consensus pattern or "chemotype" among these analogs including two hydrophobes connected by a thiourea linker.
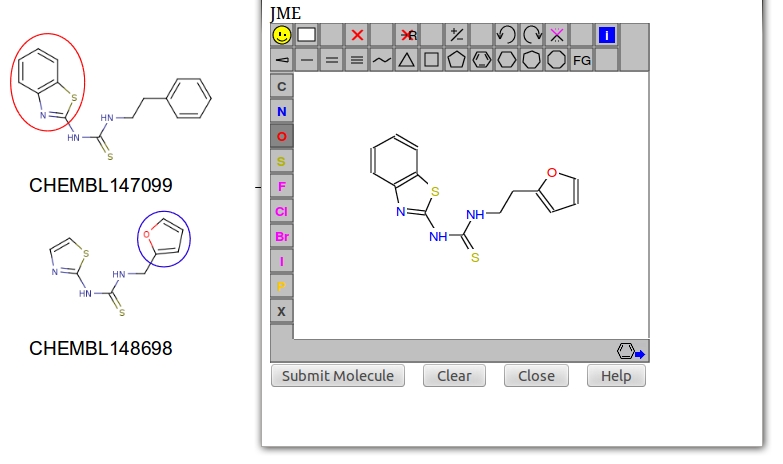
- This observed chemotype can be used to design novel HIVRT analogs by "mutating" any two analogs in a process known as "drug repurposing". Here, we use two reference compounds CHEMBL147099 and CHEMBL148698 as an example. The designed molecule is shown below.
- To confirm the designed drugs also target HIVRT, first load the original HIVRT inhibitor by clicking "load example1" in the CSNAP query page. Next, click "draw molecule," this action will open up the JME molecule editor in a seperate window. Create the designed ligands in the canvas; once completed click "submit". The JME molecule editor will automatically convert the drawn structure to SMILES string in the textarea.
- Finally, click "upload" then "submit" to preceed the calculation.
- As expected, the designed HIVRT analogs form network connectivity with the original HIVRT inhibitors with S-score=2. Again, the LTIF heatmap shows that the predicted HIVRT activity of the designed moleucle is highly specific with no off-target binding pattern.
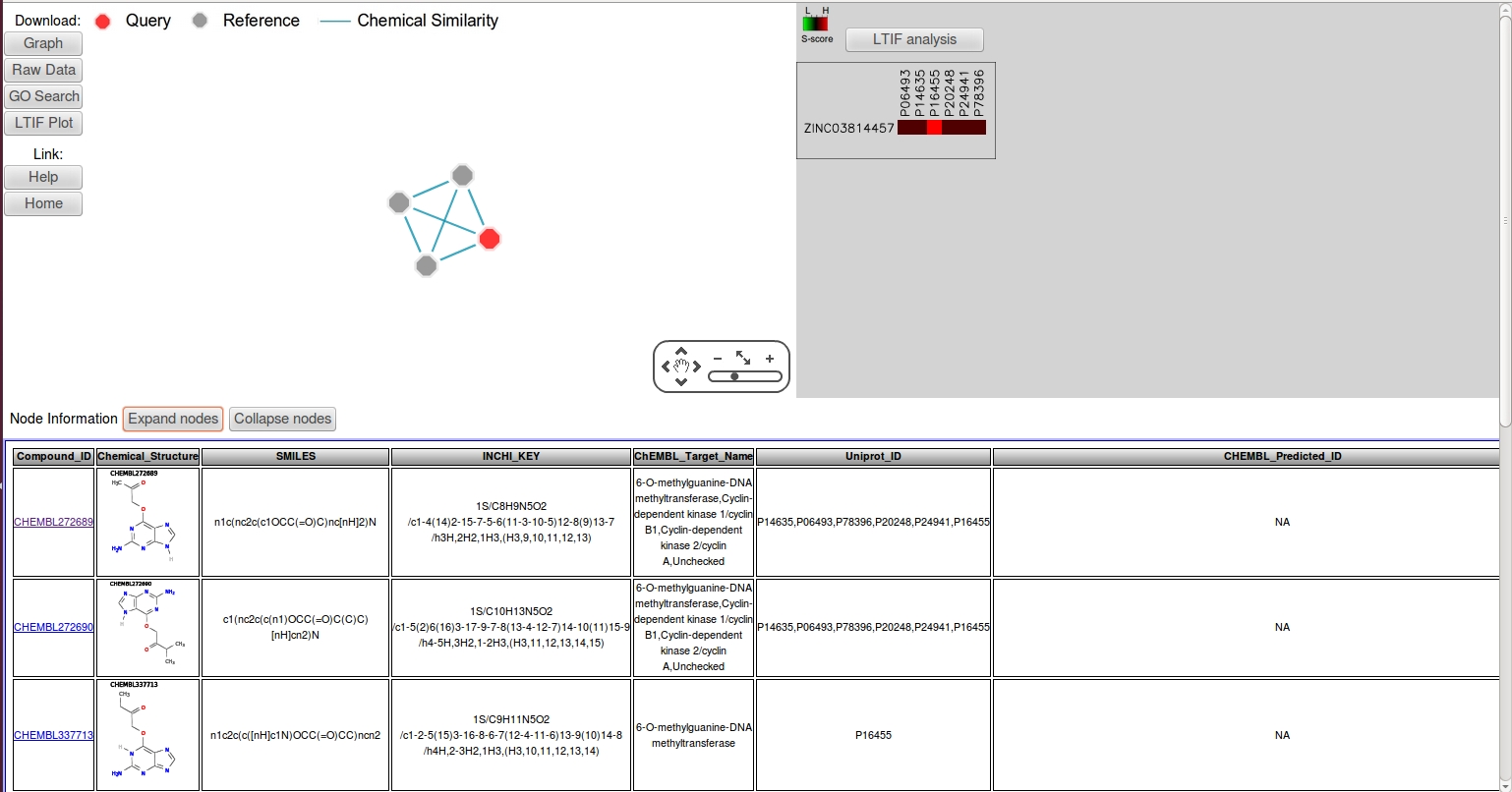
- In the input fields of CSNAP query page, click "load example2", this should load the SMILES string of a CDK2 inhibitor in the text area.
- Accept the default search parameters then click "load" then "submit" in the confirmation page.
- As shown in the LTIF panel, 4 targets have been predicted from the CSNAP analysis including CDK2 (S-score:2), and three additional targets: CDK1 (S-score:2), CCNA1 (S-score:2), CCNA2 (S-score:2), CCNB1 (S-score:2), and MGMT (S-score:3). Among the predicted off-targets, CDK1 is a known off-target for many CDK2 inhibitors due to the strucutral homology of cyclin-depedent kinases that interact with purine analogs through similar ATP catalytic pockets. Interestingly, one epigenetic target MGMT (Methylated-DNA-protein-cysteine methyltransferase) is discovered as a putative off-target.
- To gain more information about MGMT, browse the Target GO summary table by clicking the "LTIF analysis" button in the upper right panel. The GO term indicates that MGMT is inovlved in DNA damage response and repair. Thus, it is likely that the query CDK2 inhibitor may also inhibit the S phase of the cell cycle as a potential side-effect.
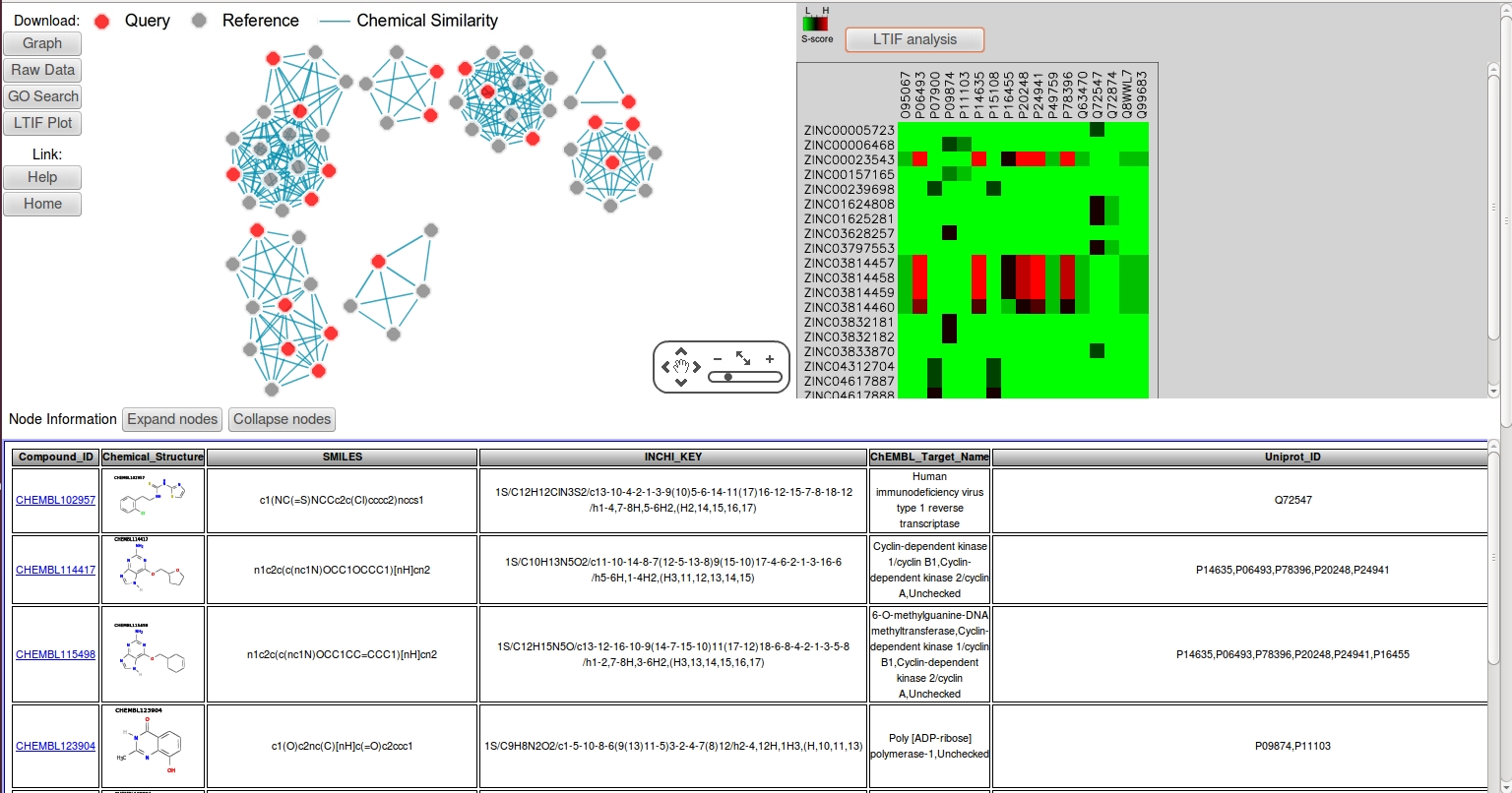
- The CSNAP server can also be applied to analyze a mixture of diverse compound structures for target prediction. For an example of this, return to the CSNAP query page and click "download example3". This will download a sdf file contains 20 compounds of 5 HSP90, HIVRT, PARP and CDK2 inhibitors. Upload the sdf file using the "upload" SDF option.
- Accept the default parameters and submit the query page as previously described.
- As shown, the pseudo-library were ordered into distinct chemical similarity networks with diferent chemotypes. The LTIF heatmap also revealed distinct target interacting patterns corresponding to different drug classes. This functionality can potentially be used to analyze structurally diverse hits from phenotypic screens.
Scenario: You have discovered an HIV inhibitor from a chemical screen that suppressed HIV replication in cell culture and would like to know the mechanism of action of this compound.
The output page is shown below:
The output page is shown below:
The output page is shown below:
Tutorial #2: Off-target Prediction of CDK2 Inhibitors
Scenario: Cyclin-dependent kinase 2 (CDK2) plays an important role in the G1/S cell cycle transition and CDK2 inhibition has been proposed as potential strategy to induce cell cycle arrest and apoptosis in the treatment of cancer. In contrast to HIVRT inhibitors from tutorial #1, many kinase inhibitors have known off-target activities due to the strong structural homology between different kinases family. Here, we will use CSNAP to identify potential off-targets of a CDK2 inhibitor.
The output page is shown below:
Tutorial #3: Target prediction and chemotype recognition from diverse compounds
The output page is shown below: