CSNAP (Chemical Similarity Network Analysis Pull-down) Web
Method Description:
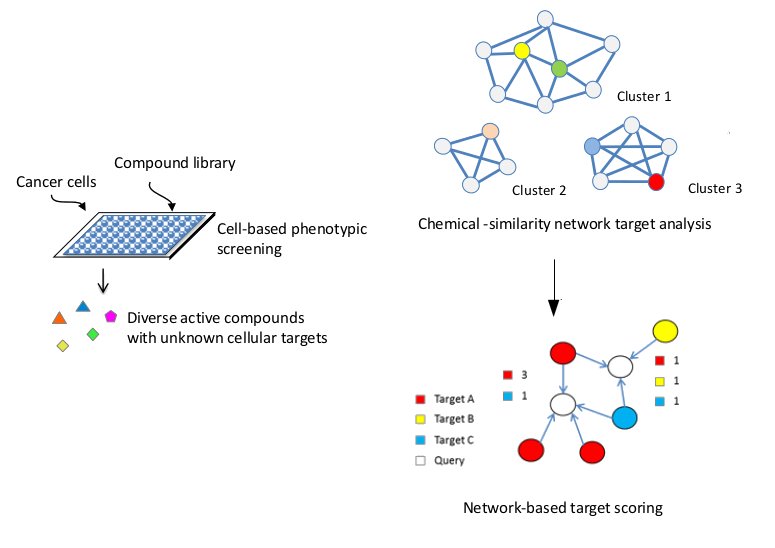
Chemical Similarity Network Analysis Pull-down (CSNAP) is a novel network-based approach for large-scale drug target identification. A simplified workflow is shown above. Diversed hit compounds retrieved from cell-based chemical screen are clustered into chemotype-based chemical similarity networks. The nearest neighbor counting functions are applied to rank the consensus targets shared among the neighbor reference nodes of each query ligand based on compound network connectivity. In contrast to conventional single-ligand inference, CSNAP can perform multi-compound target analysis and does not necessitate prior lead selection. By probing the global bioactivities of the screened hits, CSNAP aims to provide a complete profiling of compound target space to enable rapid dissection of multiple compound-target binding mechanisms and facilitate downstream lead optimization from any large phenotype-based chemical screening campaigns.
Method Validation:
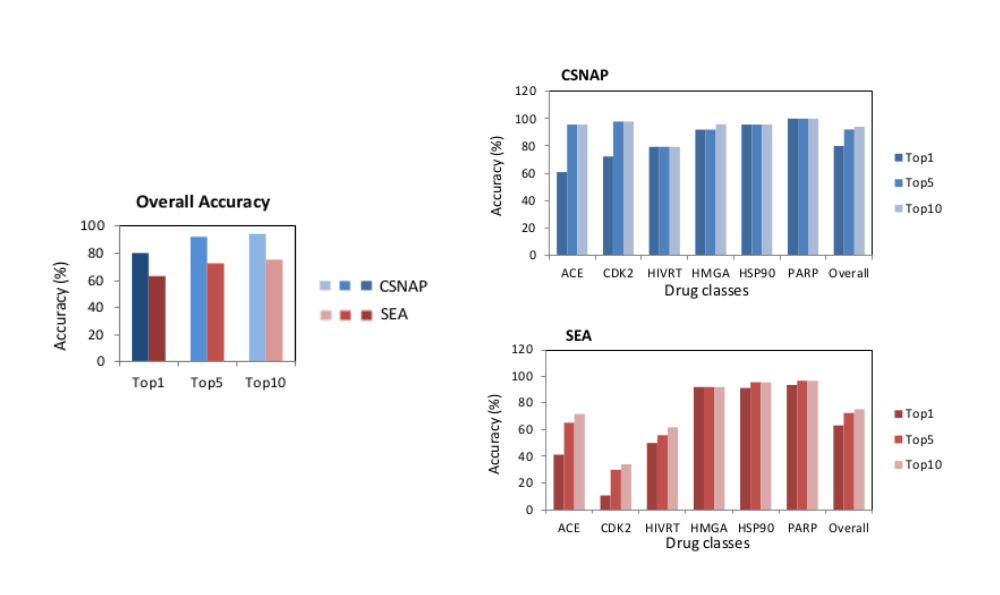
The CSNAP approach showed a substantial improvement in accuracy over the conventional ligand-based approaches for target inference. CSNAP was validated using 216 benchmark compounds set from the Directory of Useful Decoy (DUD) comprising six major compound classes. The prediction accuracy was assessed in comparison with the conventional ligand-based SEA approach.